Research
Research Concepts
In response to environmental changes, plants coordinate growth balance and metabolisms at a whole plant level including optimization of growth balance of the aerial part and root system and coordination of metabolism and morphogenesis. In order to realize the coordinated environmental responses, inter-cellular and inter-organ communication is necessary. In this communication, various signaling molecules, such as phytohormones, play important roles. However, mechanisms underlying the signal communication is not well understood yet. We are conducting research projects focusing on the role of plant hormones and other signaling molecules with focusing on nutritional responses. In particular, cytokinin, which promote plant growth and development, is an important signaling molecule in aiming improvement of plant productivity, so it is one of the main research subjects of our laboratory.
Key words:
Arabidopsis thaliana, Cytokinin, Nitrogen signaling, Phytohormones, membrane traffic
Main Research Topics:
- Elucidation of cytokinin biosynthesis and transport, and their regulatory systems
- Elucidation of mechanisms underlying organ-to-organ communication of nutritional status
- Identification of novel cytokinins and its function produced by phytopathogens
- Elucidation of the Intracellular Vesicle Transport and Meristem Size Regulation Mechanism in PATROL1
Elucidation of cytokinin biosynthesis and transport, and their regulatory systems
Cytokinins are a class of phytohormones, which enhances cytokinesis of plant cells in the presence of auxin. Over the past half-century, cytokinins have been found to be involved in various aspects of the developmental processes, including senescence, apical dominance, root proliferation, phyllotaxis, and reproductive competence. To regulate the processes, cytokinin metabolism must be tightly controlled. However, the whole picture of the cytokinin metabolic pathway has not yet been completely elucidated. The aim of our research is to fully understand cytokinin functions by elucidating cytokinin metabolism. We have identified key genes for its biosynthesis and transport (for a review, Osugi and Sakakibara 2015), and now studies regulatory mechanisms in response to environmental cues.
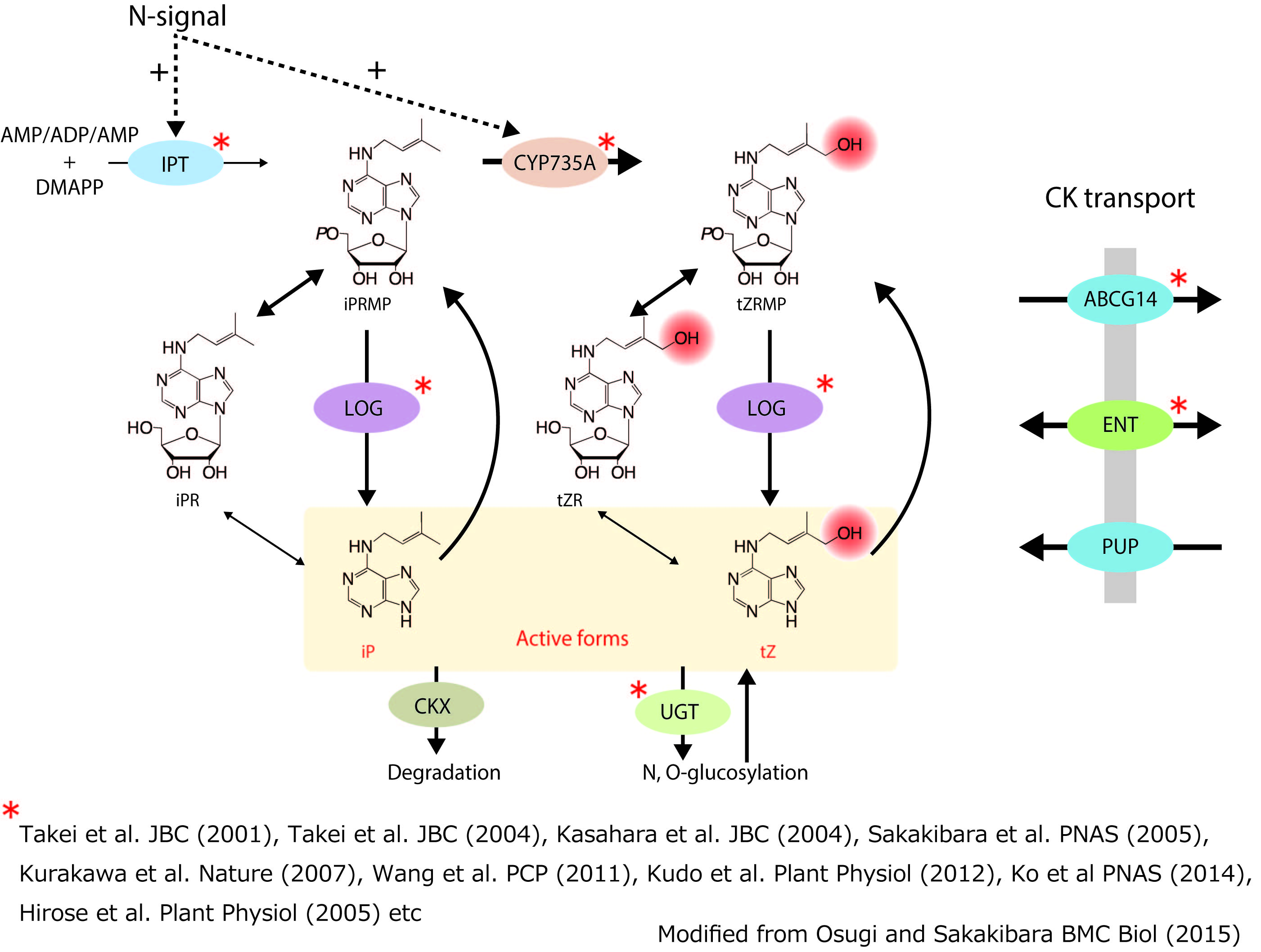
(fig. 1)Cytokinin biosynthesis pathway and transporters
Elucidation of mechanisms underlying orfile:///C:/homepae/src/en/sakakibara.html#list04gan-to-organ communication of nutritional status
Plants photosynthesize and produce all biomolecules from inorganic nutrients, but supply of inorganic nutrients from the soil is not always enough. In response to internal and external nutritional status, plants coordinately control metabolic systems and morphogenesis, and also optimize growth of the aerial part and root system at a whole plant level. In order to realize the coordinated environmental response, inter-cellular and inter-organ communication is necessary. In this communication, various signaling molecules, such as phytohormones, play important roles. We study organ-to-organ communication mechanisms in nutritional response focusing on the role of signal molecule such as phytohormones. Cytokinin is known to play an important role as an inter-organ communication molecule that is transported over long distance via xylem and phloem. We study the mechanism of long-range transport and action of cytokinins(Kiba et al. 2013; Osugi et al. 2017).
Recent studies indicate that organ-to-organ communication regulates expression of genes involved in iron absorption in response to fluctuated iron availability. We also conduct study of this molecular mechanism.
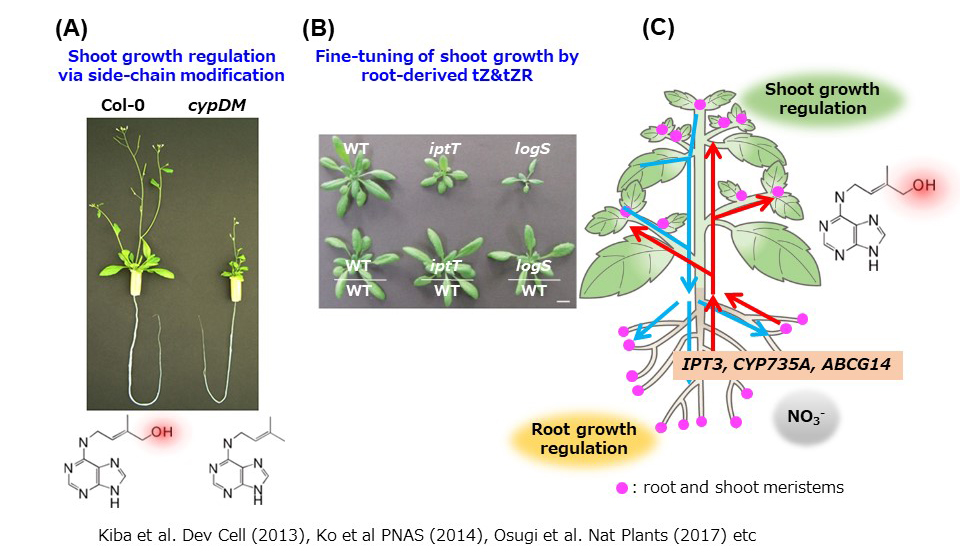
(fig. 2)Growth optimization by cytokinin via long distance transport
Identification of novel cytokinins and its function produced by phytopathogens
Cytokinin is also synthesized by phytopathogenic bacteria and a key factor in some plant diseases. For instance, A. tumefaciens infects plants and induces the formation of tumors by integrating the T-DNA region of the Ti-plasmid into the plant nuclear genome. Tumors are formed because the T-DNA encodes enzymes that modify the synthesis of two plant growth hormones, auxin and cytokinin. We had demonstrated that a cytokinin biosynthesis enzyme, Tmr, which is encoded by the Agrobacterium T-DNA region, is targeted to plastids of infected plant cells, and increases cytokinin production by modifying the biosynthetic pathway in the host plant(Sakakibara et al. 2005; Ueda et al. 2012).
As another example, Rhodococcus fascians infection causes unique leafy gall symptoms reminiscent of cytokinin over-effect. The fasciation (fas) locus, an operon encoding several genes homologous to cytokinin biosynthesis and metabolism. However, real function of these genes has not been fully elucidated. We are tackling research to identify novel cytokinin species and its function.
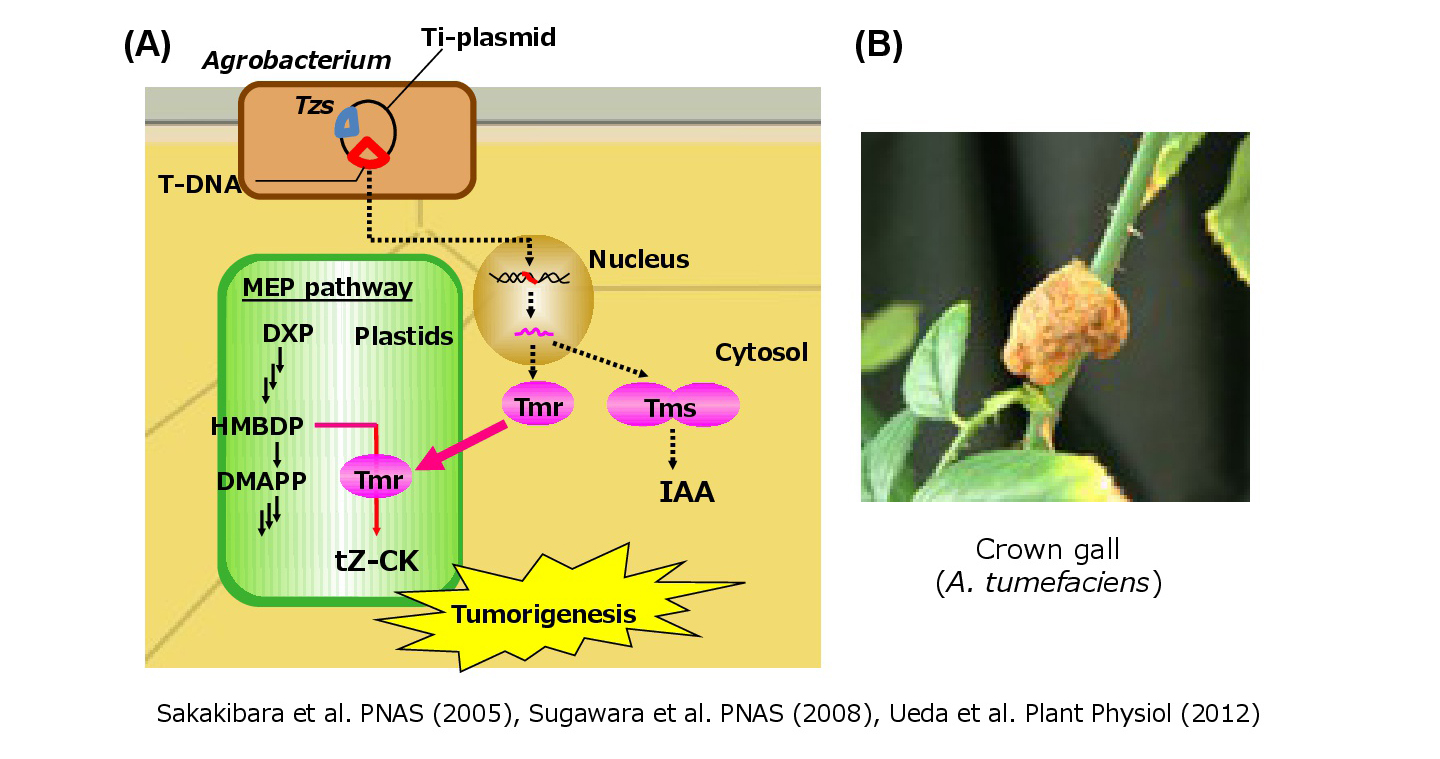
(fig. 3)Agrobacterium increases cytokinin production in the host plant.
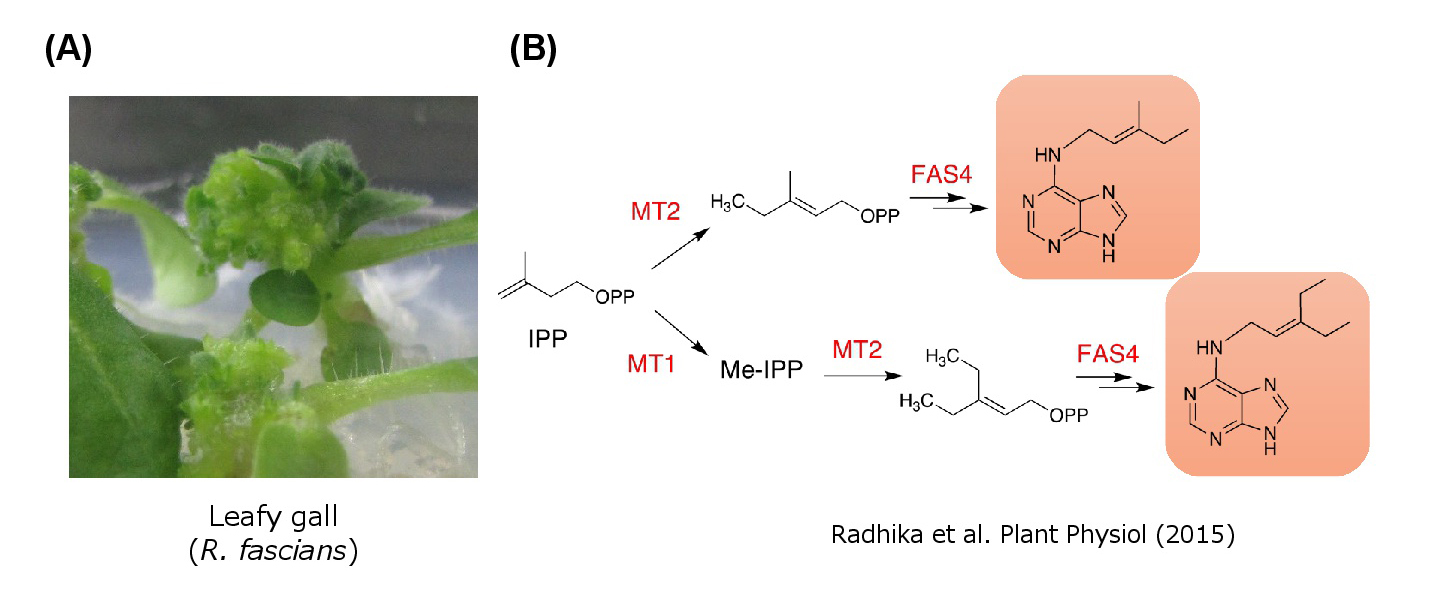
(fig. 4) Rhodococcus fascians infection causes unique leafy gall symptoms reminiscent of cytokinin over-effect.
4. Membrane traffic and meristem size regulation mechanism in PATROL1

(Figure 5) Discovery of the patrol1 (proton-ATPase control 1) mutant and overview of functional analysis of the PATROL1 gene
(A) Development of a system to monitor stomatal aperture changes in response to CO2 concentration using thermography(B) Isolation of CO2-response stomatal mutants (high leaf temperature 1, 2)
(C) The roots of the patrol1 mutant (ht2) are short
(D) PATROL1 is a protein containing a MUN domain
(E) In animals, the MUN domain is involved in intracellular vesicle transport and promotes SNARE complex formation
(F) The vesicle transport targets of PATROL1 differ between guard cells and roots
Stomata exist on the surface of plant leaves and serve as gateways for gas exchange. Since the stomatal aperture changes in response to CO2 concentration, it was hypothesized that plants possess CO2 sensors. When stomata open, evaporative cooling via transpiration lowers leaf temperature. We indirectly monitored stomatal aperture using thermography and successfully isolated the first mutants involved in the stomatal CO2 response (Hashimoto et al., Nat. Cell Biol. 2006; Hashimoto-Sugimoto et al., Nat. Commun. 2013; Fig. 6A, B). The first mutant we isolated was ht1 (high leaf temperature 1), whose causal gene HT1 encodes a kinase. We found that its activity correlated with CO2 responsiveness and that it functions as a key CO2-specific enzyme (Hashimoto et al., Nat. Cell Biol. 2006; Hashimoto-Sugimoto et al., J. Exp. Bot. 2016).
Meanwhile, the ht2 mutant (later renamed patrol1) exhibited impaired stomatal opening and reduced overall plant size (Hashimoto-Sugimoto et al., Nat. Commun. 2013; Higaki et al., Plant Cell Physiol. 2014; Fig. 6C). The PATROL1 gene, responsible for this phenotype, was predicted to encode a protein containing a MUN domain, which is widely conserved across organisms (Fig. 6D). In animals, proteins such as Munc13 and unc13, which have MUN domains, are essential for docking secretory vesicles containing neurotransmitters to the plasma membrane. The MUN domain promotes the formation of the SNARE complex (Fig. 6E). In the patrol1 mutant, the amount of the H+-ATPase (AHA1), which drives stomatal opening, was reduced in the guard cell plasma membrane. This suggested that PATROL1 promotes the localization of AHA1 to the guard cell plasma membrane (Hashimoto-Sugimoto et al., Nat. Commun. 2013).
PATROL1 is also strongly expressed in roots, and the mutant shows a reduced meristem size (Notaguchi et al., Planta 2024). Since H+-ATPases are involved in cell elongation, it was hypothesized that AHA1 levels might also be altered in root plasma membranes. However, the amount of AHA1 was unchanged compared to wild-type, and even AHA2, the major isoform in roots, showed no difference in abundance (Notaguchi et al., Planta 2024; Fig. 6F). Thus, we are currently working to elucidate the molecular mechanism by which PATROL1 regulates meristem size.
Because PATROL1 influences biomass production (Sugimoto-Hashimoto et al., Nat. Commun. 2013; Kimura et al., J. Exp. Bot. 2020), elucidating the molecular mechanisms of PATROL1-mediated intracellular vesicle transport could contribute to the development of molecular breeding strategies for high-yield crops and environmentally beneficial greening.
